Bioinformatics alerts for viral variant impacts on molecular tests
Posted on Wednesday, August 11, 2021
By
Topic: What is Trending in Science

Bioinformatics tools helped developers kick start molecular diagnostics tests for COVID-19, and bioinformatics tools can also help us learn how to react faster to disease outbreaks. That’s what I learned from interviewing NEB scientist Brad Langhorst. As SARS-CoV-2 variants spread, the primers used for diagnostic tests must be continuously verified. NEB’s open access Primer Monitor tool makes it easier for scientists and clinicians to do just that.
What prompted NEB to develop the Primer Monitor software?
The major motivation for building this tool was the number of requests we received while interacting with other scientists working on SARS-CoV-2 diagnostic testing as they prepared their applications to the federal government for Emergency Use Authorization.
These scientists needed information to show how robust their primer sets were going to be to genetic variation. Would their primers continue to work with COVID variants? What would happen if the primers interacted with other DNA, such as human background DNA or other pathogens? At that point, I was fielding these questions and handling them one by one with a custom analysis, but researchers needed software to evaluate their test primer sets and be automatically updated as conditions changed worldwide.
What is the Primer Monitor?
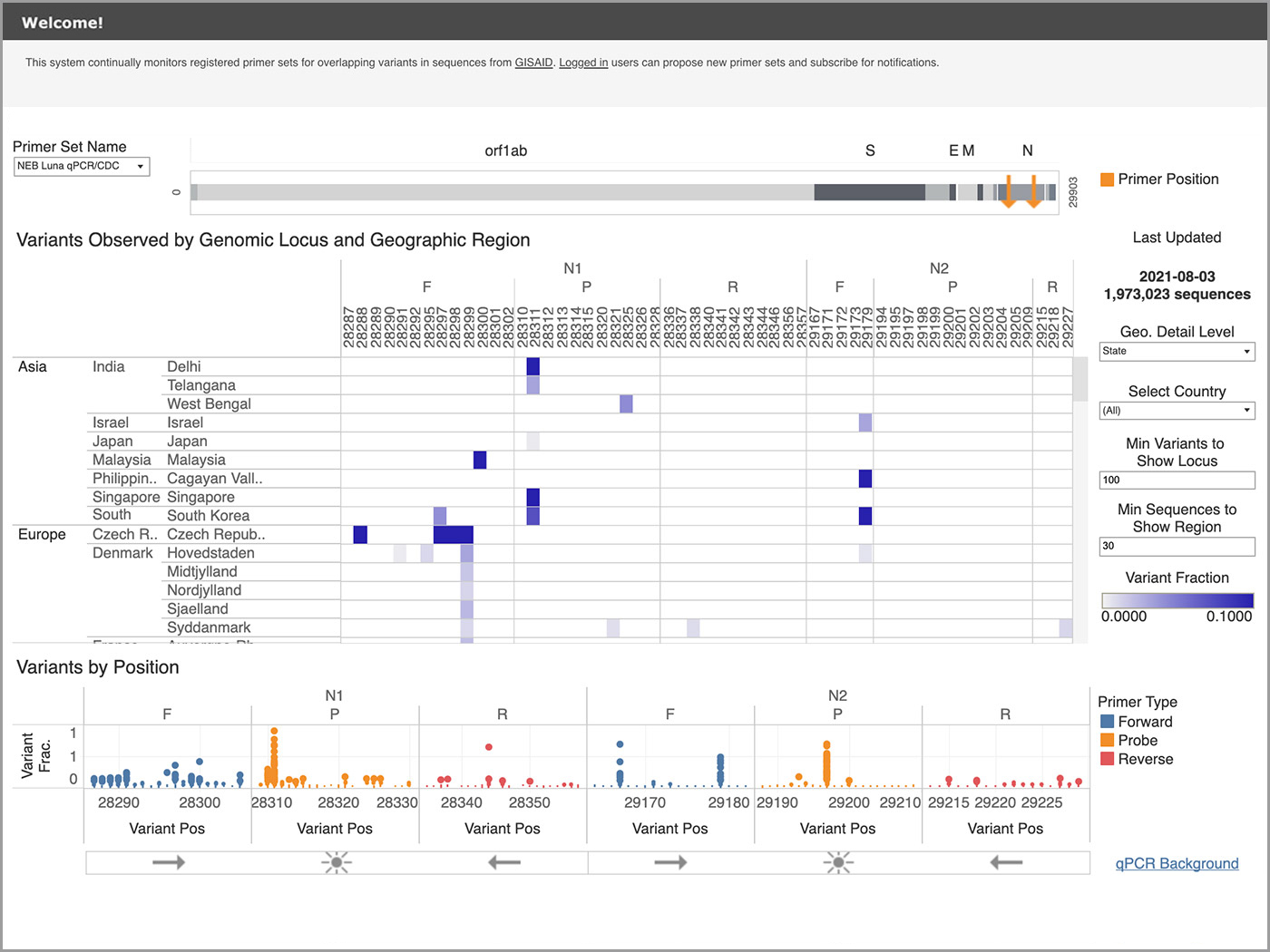
The Primer Monitor is an open database, visualization tool and notification system developed by New England Biolabs and updated with global public health data from GISAID (a global initiative that promotes the sharing of data from SARS-CoV-2 and influenza viruses). This bioinformatics tool has a website solely dedicated to helping users understand how variants interact with molecular diagnostics detection test primers.
What is the risk to public health if the performance of SARS-CoV-2 molecular diagnostic tests slip?
From a technical perspective, why has the FDA required SARS-CoV-2 test developers to monitor for viral variant impacts?
Are some tests at greater risk for failure against viral variants?
Does a change in test sensitivity with SARS-CoV-2 variants matter?
It absolutely matters. The more sensitive the test is, the bigger the window you have to detect the virus. If there is a significant change in test sensitivity for a certain SARS-CoV-2 variant, tests conducted earlier or later during the course of the infection might not capture what's really happening and lead to a false negatives. That’s a serious concern when someone is contagious and in contact with family members, or the community.
The fact that you can subscribe to a primer set and automatically be notified down the road to changes in the variant landscape for a given assay is very useful.
What is the most powerful feature of the Primer Monitor?
The subscription alerts feature. Scientists are busy. The fact that you can subscribe to a primer set and automatically be notified down the road to changes in the variant landscape for a given assay is very useful.
I will add that the geographic specificity in the Primer Monitor is another compelling feature. Even though lineages of the virus have spread dramatically around the world, there is still a geography component to outbreaks. It's helpful to know where a test isn’t working due to the prevalence of a given mutation in viral genomes spreading in that region. The Primer Monitor provides those insights thanks to the detailed granularity provided in the viral sequence data.
Is that how you would define a viral hotspot?
The term hotspot has been used in a couple of ways. We have heard it used geographically, but it’s also used in bioinformatics to refer to a specific area of the genome prone to variation. A nexus might be a better term for what we have observed several times worldwide using the Primer Monitor. A nexus is when a particular viral variant is connected to a geographical region that uses a test with variant impacts to its primer set. The Primer Monitor showed one that first arose in Pueblo, Mexico, and then another in South Korea where it was problematic for one particular assay to maintain robustness.
Why is sharing data on primer performance for variants in the Primer Monitor helpful to science?
There is so much to gain from democratizing access to data. Viral variant impact on testing is an area where the researchers who are willing to share can benefit others. Most molecular diagnostics tests are using the same or similar areas for priming. If an impact emerges, that becomes a shared response. Using the Primer Monitor fits a pandemic trend that we’re seeing – where walls erected for scientists to work quietly on problems until they are ready to share – have been broken down. The goal is for data in the Primer Monitor to enable scientists to move faster together to correct any impacts.
Broadly, how has the field of bioinformatics supported viral detection during the pandemic?
Are healthcare providers potential users of the Primer Monitor tool?
Doctors and labs relying on tests could use Primer Monitor to learn, for example, that an assay could be problematic in 30% of cases in their location. That would be a good prompt to cross-check results with a secondary orthogonal assay. It’s been great that clinicians and other public health personnel have been much more deeply involved in sequencing and the evolution of diagnostics to fight the pandemic.
What is the GISAID, and how was it used before COVID-19?
What other bioinformatics investigations could scientists use the Primer Monitor tool for?
Information about how primers interact with sequences and how variants impact tests could help to refine our understanding of the "rules" for multiplex amplification reactions. It would be helpful to predict that a variant will impact a primer pair with a two-fold reduction in efficiency, but I don't think that's possible with our current models. GISAID provides consensus sequences rather than raw data; however, with enough consensus data I think we can refine those rules to make better predictions. Many groups are also submitting raw reads to sequence repositories like the SRA hosted by NCBI, which can further help with this challenge.
What would you like to learn from scientists using the Primer Monitor that could help improve the tool?
What about other diseases? Can those impact COVID-19 molecular diagnostics tests?
Right. Another common question from scientists I spoke with was, "Do these primers hit other organisms?" That is an additional bioinformatics challenge for when we are asking, "Is this going to be specific to SARS-CoV-2, or not?" The FDA has a panel of organisms – that you might find in someone’s microbiome. Developers need to confirm that their test primers are unlikely to amplify any of these species. You don't want false positives from other organisms, whether they are disease-causing or not. Primer Monitor doesn't handle that challenge right now, but might at some point in the future if there is enough need for it.
Are there any bioinformatics lessons from the pandemic that can be applied to how we test for other infectious disease outbreaks, like influenza or even zoonic disease outbreaks in wildlife or livestock?
COVID-19 has expanded testing infrastructure, and we can leverage that for the greater good. Variant outbreaks during this pandemic have been frustrating, and preemptive planning based on existing infrastructure was difficult. We were more prepared that we could have been because past disease outbreaks, such as the Zika virus outbreak, were essentially ‘training’ for several sequencing groups who made important contributions to SARS-CoV-2. Both Zika and Ebola were major lessons on what can be done in the field. Going forward, I hope that bioinformatics analysis will continue to help human society learn to become more organized and able to respond faster.
Any parting words of caution on monitoring viral genetic drift for impacts to molecular diagnostics?
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.