Cellular Analysis
Choose Type:
-
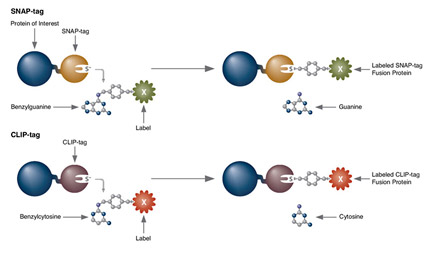
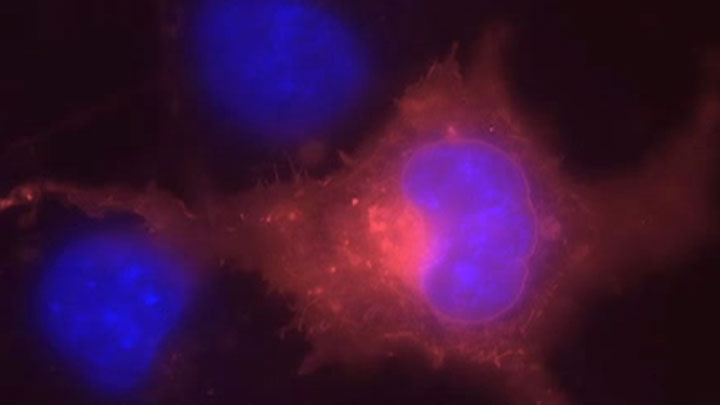
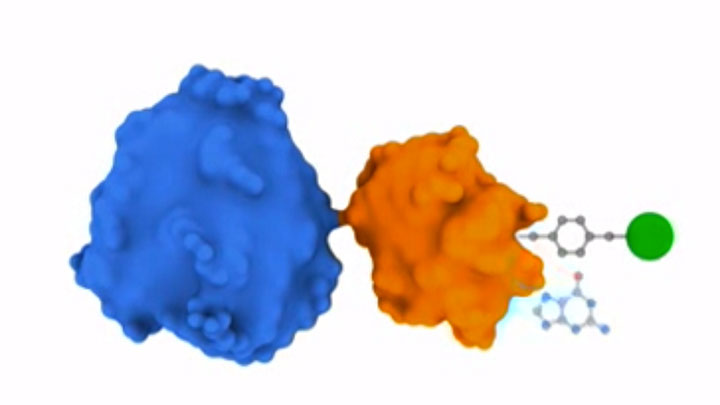
SNAP-tag® Technologies: Tools to Study Protein Function
Read about the NEB’s set of protein tools for the specific labeling (SNAP-, CLIP-, ACP- and MCP-tags) of fusion proteins.
- Cellular Imaging & Analysis Brochure
- Purification Beads, Columns and Resins Brochure
- Building Blocks
- Comparison of SNAP-tag®/CLIP-tag™ Technologies to GFP
- SNAP-tag® and CLIP-tag™ Substrate Selection Chart
- SNAP-tag®/CLIP-tag® Cloning Vector Selection Chart
- Labeling with SNAP-tag® Technology Troubleshooting Guide
- Genome-wide profiling of nuclease protected domains reveals physical properties of chromatin (2019)
- In Vitro Reconstitution of Thermococcus Species 9°N Okazaki Fragment Maturation (2015)
Feature Articles
Brochures
Selection Tools
Troubleshooting Guides
Posters
- Dellagiacoma, C. et al. (2010) Targeted photoswitchable probe for nanoscopy of biological structures Chembiochem; PubMedID: 20540058, DOI: 10.1002/Cbic.201000189
- Hein B. et al. (2010) Stimulated emission depletion nanoscopy of living cells using SNAP-Tag fusion proteins Biophys J; 98 , 158-163 . PubMedID: 20074516
- Rhee S. G. et al. (2010) Methods for detection and measurement of hydrogen peroxide inside and outside of cells Mol Cell; 29 , 539-549 . PubMedID: 20526816
- Alvarez-Curto J. et al. (2010) Ligand regulation of the quaternary organization of cell surface M3 muscarinic acetylcholine receptors analyzed by fluorescence resonance energy transfer (FRET) imaging and homogenous time-resolved FRET J Biol Chem; 285 , 23318-23330 . PubMedID: 20489201
- Geissbuehler M. et al. (2010) Triplet imaging of oxygen consumption during the contraction of a single smooth muscle cell Biophys J; 98 , 339-349 . PubMedID: 22259112
- Kampmeier, F. et al. (2010) Rapid optical imaging of EGF receptor expression with a single-chain antibody SNAP-tag fusion protein Eur J Nucl Med Mol Imaging; PubMedID: 20449589, DOI: 10.007/S00259-010-1482-5
- Waichman S. et al. (2010) Functional Immobilization and Patterning of Proteins by an Enzymatic Transfer Reaction Anal Chem; 82 , 1478-1485 . PubMedID: 20092261
- Zelman-Femiak, M. et al. (2010) Covalent quantum dot receptor linkage via the acyl carrier protein for single-molecule tracking, internalization, and trafficking studies Biotechniques; 49, 2. PubMedID: 20701592
- Mosiewicz, K. A. et al. (2010) Phosphopantetheinyl Transferase-Catalyzed Formation of Bioactive Hydrogels for Tissue Engineering J Am Chem Soc; 132, 5972-5974 . PubMedID: 20373804
- Campos, C. et al. (2010) Labeling cell structures and tracking cell lineage in zebrafish using SNAP-Tag Dev Dyn; 240 , 820-827. PubMedID: 21360787
- Srikun, D. et al. (2010) Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-tag protein labeling J Am Chem Soc; 132 , 4455-4465 . PubMedID: 20201528
- Ciruela F. et al. (2010) Lighting up multiprotein complexes: lessons from GPCR oligomerization Trends Biotechnol; 28, 407-415 . PubMedID: 20542584
- Kamiya M. and Johnsson K. (2010) Localizable and Highly Sensitive Calcium Indicator Based on a BODIPY Fluorophore Anal Chem; 82 , 6472-6479 . PubMedID: 20590099
- Jansen L. et al (2007) Propagation of centromeric chromatin requires exit from mitosis J Cell Biol; 176, 795-805. PubMedID: 17339380
- Böhme. et al. (2007) Tracking of human Y receptors in living cells- A fluorescence approach Peptides; 28, 226-234 . PubMedID: 17207557
- Lemercier, G. et al. (2007) Inducing and sensing protein-protein interactions in living cells by selective cross-linking Angew Chem Int Ed Eng; 4281-4284 . PubMedID: 17465435
- O'Hare H.M. et al. (2007) Chemical probes shed light on protein function Curr Opin Struct Biol; 17 , 488-94 . PubMedID: 17851069
- Johnsson N. and Johnsson K. (2007) Chemical tools for biomolecular imaging ACS Chem Biol; 2 , 31-38 . PubMedID: 17243781
- Stein, V. et al. (2007) A covalent chemical genotype-phenotype linkage for in vitro protein evolution Chembiochem; 8, 2191-4 . PubMedID: 17948318
- Liu E and Bruner S. D. (2007) Rational manipulation of carrier-domain geometry in nonribosomal peptide synthetases Chembiochem; 8, 617 - 621 . PubMedID: 17335097
- George N. et al. (2004) Specific labeling of cell surface proteins with chemically diverse compounds J Am Chem Soc; 126, 8896-8897 . PubMedID: 15264811
- Kindermann M. et al. (2004) Synthesis and characterization of bifunctional probes for the specific labeling of fusion proteins Bioorg Med Chem Lett; 14, 2725-2728 .
- Huber W. et al. (2004) SPR-based interaction studies with small molecular weight ligands using hAGT fusion proteins Anal Biochem; 333, 280-288 . PubMedID: 15450803
- Keppler A. et al. (2004) Labeling of fusion proteins of O6-alkylguanine-DNA alkyltransferase with small molecules in vitro and in vivo Methods; 32, 437-444. PubMedID: 15003606
- Keppler A. et al. (2004) Labeling of fusion proteins with synthetic fluorophores in live cells Proc Natl Acad Sci U S A; 101, 9955-9959.
- La Clair, J.J. et al. (2004) Manipulation of carrier proteins in antibiotic biosynthesis Chem Biol; 11, 195-201 . PubMedID: 15123281
- Damoiseaux, R. et al (2002) Towards the generation of artificial O6-alkylguanine-DNA alkyltransferases: in vitro selection of antibodies with reactive cysteine residues Chembiochem; 3, 573-575 . PubMedID: 12325014
- Fururta, K. et al. (2008) Diffusion and directed movement: in vitro motile properties of fission yeast kinesin-14 Plk1 J Biol Chem; 283 , 36465-36473 . PubMedID: 18984586
- Schulz C. and Köhn M. (2008) Simultaneous protein tagging in two colors Chem Biol; 15, PubMedID: 18291310
- Mao S. et al. (2008) Optical lock-in detection of FRET using synthetic and genetically encoded optical switches Biophys J; 94, 4515-24 . PubMedID: 18281383
- Erhardt, S. et al. (2008) Genome-wide analysis reveals a cell cycle-dependent mechanism controling centromere propagation J Cell Biol; 183 , 805-818 . PubMedID: 19047461
- Generosi J. et al. (2008) AMPA receptor imaging by infrared scanning near-field optical microscopy Physica Status Solidi C: Current Topics in Solid State Physics; 5, 2641-2644 .
- Southwell, A.L. et al. (2008) Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity J Neurosci; 28, 9013-20 . PubMedID: 18768695
- Kropf M. et al. (2008) Subunit-specific surface mobility of differentially labeled AMPA receptor subunits Eur J Cell Biol; 87, 763-778 . PubMedID: 18547676
- Chidley C. et al. (2008) A designed protein for the specific and covalent heteroconjugation of biomolecules Bioconjugate Chem; 19 , 1753-1756 . PubMedID: 18754573
- Maurel D. et al. (2008) Cell-surface protein-protein interaction analysis with time-resolved FRET and SNAP-tag technologies: application to GPCR oligomerization Nat Methods; 5, 561-7 . PubMedID: 18488035
- Generosi J. et al. (2008) Photobleaching-free infrared near-field microscopy localizes molecules in neurons J Appl Physiol; 104, 106102-1/3.
- Tomat, E. et al. (2008) Organelle-specific zinc detection using zinpyr-labeled fusion proteins in live cells J Am Chem Soc; 130 , PubMedID: 18973293
- Gautier A. et al. (2008) An engineered protein tag for multiprotein labeling in living cells Chem Biol; 15, 128-136. PubMedID: 18291317
- Eckhardt, M. et al. (2011) A SNAP-tagged detivative of HIV-1 - A versatile tool to study virus-cell interactions PLoS One; 6:e22007 . PubMedID: 21799764, DOI: 10.137/journal. P One .0022007
- Hoskins, A. et al. (2011) Ordered and dynamic assembly of single spliceoseoms Science; 331 , 1289 . PubMedID: 21393538
- Zhou Z. et al. (2007) Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases ACS Chem Biol; 2, 337-346 . PubMedID: 17465518
- Stenoien D. L. et al. (2007) Cellular trafficking of phospholamban and formation of functional sarcoplasmic reticulum during myocyte differentiation Am J Physiol Cell Physiol; 292 , C2084-C2094 . PubMedID: 17287364
- Pick H. et al. (2007) Distribution plasticity of the human estrogen receptor alpha in live cells: distinct imaging of consecutively expressed receptors J Mol Biol; 14, 1213-1223. PubMedID: 17991486
- Mottram L. F. et al. (2007) A Concise Synthesis of the Pennsylvania green fluorophore and labeling of intracellular targets with O6-Benzylguanine Derivatives Org Lett; 9, 3741-3744 . PubMedID: 17705395
- Gendreizig S. et al. (2003) Covalent labeling of fusion proteins with chemical probes in living cells Chimia; 57, 181-183 .
- Juillerat A. et al. (2003) Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo Chem Biol; 10, 313-317 .
- Gendreizig, S. et al. (2003) Induced protein dimerizaton in vivo through covalent labeling J Am Chem Soc; 125, 14970-14971 . PubMedID: 14653715
- Keppler A. et al. (2003) A general method for the covalent labeling of fusion proteins with small molecules in vivo Nat Biotechnol; 21, 86-89 .
- Kindermann M. et al. (2003) Covalent and selective immobilization of fusion proteins J Am Chem Soc; 125, 7810-7811 . PubMedID: 12822993
- Iversen L. et al. (2008) Templated protein assembly on micro-contact-printed surface patterns. Use of the SNAP-tag protein functionality Langmuir; May 17, PubMedID: 18484753
- Howland S.W. et al. (2008) Inducing efficient cross-priming using antigen-coated yeast particles J Immunother ; 31 , 607-19. PubMedID: 18600183
- Gautier A. et al. (2008) AGT/SNAP-Tag: A versatile tag for covalent protein labeling from probes and tags to study biomolecular function Ed. Edited by Miller, L. W. ; 89-107 .
- Sunbul M. et al. (2008) Enzyme catalyzed site-specific protein labeling and cell imaging with quantum dots Chem Commun; 5927-5929 . PubMedID: 19030541
- Adams D. G. et al. (2008) Cellular Ser/Thr-kinase assays using generic peptide substrates Curr Chem Genomics; 1 , 54-64 . PubMedID: 20161828
- Johnson K. (2008) SNAP-tag Technologies: Novel tools to study protein function NEB Expressions ; 3.3 , 1-3 .
- McMurray, M.A. and Thorner, J. (2008) Septin stability and recycling during dynamic structural transitions in cell division and development Curr Biol; 18 , 1203-1208 . PubMedID: 18701287
- Banala J. et al. (2008) Caged substrates for protein labeling and immobilization Chembiochem; 4, PubMedID: 18033718
- Lin M.Z. and Wang L. (2008) Selective labeling of proteins with chemical probes in living cells Physiology; 23 , 131-141 . PubMedID: 18556466
- Engin S. et al. (2010) Benzylguanine Thiol self-assembled monolayers for the immobilization of SNAP-tag proteins on microcontact-printed surface structures Langmuir; ASAP, PubMedID: 20369837
- Maurel D. et al. (2010) Photoactivatable and photoconvertible fluorescent probes for protein labeling ACS Chem Biol; PubMedID: 20218675
- Nicolle O. et al. (2010) Development of SNAP-tag-mediated live cell labeling as an alternative to GFP in Porphyromonas gingivalis FEMS�Immunol�Med Microbiol; 59 , 357-363 . PubMedID: 20482622
- Ruggiu A. A. et al. (2010) Fura-2FF-based calcium indicator for protein labeling Org Biomol Chem; 8 , 3398-3401 . PubMedID: 20556282
- Jongsma M.A., Litjens R. H. (2006) Self-assembling protein arrays on DNA chips by auto-labeling fusion proteins with a single DNA address Proteomics; 6, 2650-2655 . PubMedID: 16596705
- Keppler A. et al. (2006) Fluorophores for live cell imaging of AGT fusion proteins across the visible spectrum Biotechniques; 41, 167-75 . PubMedID: 16925018
- Prummer M. et al. (2006) Post-translational covalent labeling reveals heterogeneous mobility of individual G protein-coupled receptors in living cells Chembiochem; 7, 908-911 . PubMedID: 16607667
- Meyer B.H. et al. (2006) Covalent labeling of cell-surface proteins for in vivo FRET studies FEBS Lett; 580, 1654-1658 . PubMedID: 16497304
- Sielaff I. et al. (2006) Protein function microarrays based on self-immobilizing and self-labeling fusion proteins Chembiochem; 7, 194-202. PubMedID: 16342318
- Heinis C. et al. (2006) Evolving the substrate specificity of O6 alkylguanine DNA alkyltransferase through loop insertion for applications in molecular imaging ACS Chem Biol; 1, 575-584. PubMedID: 17168553
- Gronemeyer T. et al. (2006) Adding value to fusion proteins through covalent labeling Curr Opin Biotechnol; 16 , PubMedID: 15967656
- Tirat A. et al. (2006) Evaluation of two novel tag-based labeling technologies for site-specific modification of proteins Int J Biol Macromol.; 39, 66-76. PubMedID: 16503347
- Jacquier V. et al. (2006) Visualizing receptor trafficking in living Proc Natl Acad Sci U S A; 103, 14325-14330 . PubMedID: 16980412
- Gronemeyer T. et al. (2006) Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling Prot Eng Des Sel; 19, 309-16 . PubMedID: 12725859
- Krayl M. et al. (2006) Fluorescence-mediated analysis of mitochondrial preprotein import in vitro Anal Biochem; 335, 81-9. PubMedID: 16750157
- Meyer B.H. et al. (2006) FRET imaging reveals that functional neurokinin-1 receptors are monomeric and reside in membrane microdomains of live cells Proc Natl Acad Sci U S A; 103, 2138-43 . PubMedID: 16461466
- Kufer S.K. et al. (2005) Covalent immobilization of recombinant fusion proteins with hAGT for single molecule force spectroscopy Eur Biophys J; 35, 72-78. PubMedID: 16160825
- Cravatt B.F. (2005) Live chemical reports from the cell surface Chem Biol; 12, 954-956 . PubMedID: 16183017
- Vivero-Pol L. et al. (2005) Multicolor imaging of cell surface proteins J Am Chem Soc; 127, 12770-12771 . PubMedID: 16159249
- Juillerat A. et al. (2005) Engineering substrate specificity of O6-alkylguanine-DNA alkyltransferase for specific protein labeling in living cells Chembiochem; 6, 1263-1269 . PubMedID: 15934048
- Yin J. et al. (2005) Labeling proteins with small molecules by site-specific posttranslational modification J Am Chem Soc; 126 , 7754-7755 . PubMedID: 15212504
- Johnsson N. et al. (2005) Protein chemistry on the surface of living cells Chembiochem; 6 , 47-52 . PubMedID: 15558647
- Yin J. et al. (2005) Single-cell FRET imaging of transferrin receptor trafficking dynamics by Sfp-catalyzed, site-specific protein labeling Chem Biol; 12, 999-1006 . PubMedID: 16183024
- Tugulu S. et al. (2005) Protein-functionalized polymer brushes Biomacromolecules; 6, 1602-1607. PubMedID: 15877383
- Regoes A. et al. (2005) SNAP-tag mediated live cell labeling as an alternative to GFP in anaerobic organisms Biotechniques; 39, 809-812 .
- Foltz D.R. et al. (2009) Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP Cell; 137 , 472-84 . PubMedID: 19410544
- Milenkovic L. et al. (2009) Lateral transport of smoothened from the plasma membrane to the membrane of the cilium J Cell Biol; 187 , 365-374 . PubMedID: 19193035
- Böhme I and Beck-Sickinger A. G. (2009) Illuminating the life of GPCRs Cell Commun Signal; 7 , 16 . PubMedID: 19602276
- Tivari R. and Parang K. (2009) Protein conjugates of SH3-domain ligands and ATP- competitive inhibitors as bivalent inhibitors of protein kinases Chembiochem; 10, 2445 - 2448 . PubMedID: 19731277
- Chattopadhaya S. et al. (2009) Expanding the chemical Biologist's tool kit: chemical labelling strategies and its applications Curr Med Chem; 16 , 4527-4543 . PubMedID: 19903152
- Neugart F. et al. (2009) Detection of ligand-induced CNTF receptor dimers in living cells by fluorescence cross correlation spectroscopy Biochim Biophys Acta; 1788 , 1890-1900 . PubMedID: 19482006
- Farr G. A. et al. (2009) Membrane proteins follow multiple pathways to the basolateral cell surface in polarized epithelial cells J Cell Biol; 186 , 269-282 . PubMedID: 19620635
- Brun M.A. et al. (2009) Semisynthetic fluorescent sensor proteins based on self-labeling protein tags J Am Chem Soc; 131 , 5873-5784 . PubMedID: 19348459
- Bannwarth et. al. (2009) Indo-1 Derivatives for local calcium sensing ACS Chem Biol; 4 , 179-190 . PubMedID: 19193035
- Ahier A. et al. (2009) A new family of receptor tyrosine kinases with a venus flytrap binding domain in insects and other invertebrates activated by aminoacids PLoS One; 4, e5651 . PubMedID: 19461966
- Sletten E. and Bertozzi C. (2009) Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality Angew Chem Int Ed Eng; 48 , 6974-6998 . PubMedID: 19714693
- Eggeling C. et al. (2009) Direct observation of the nanoscale dynamics of membrane lipids in a living cell Nature; 457 , 1159-1163. PubMedID: 19098897
- Uano Y. and Matsuzaki K. (2009) Tag-probe labeling methods for live-cell imaging of membrane proteins Biochim Biophys Acta; 1788 , 2124-2131 . PubMedID: 19646952
- Degorce F. et al. (2009) HTRF: A technology tailored for drug discovery - a review of theoretical aspects and recent applications Curr Chem Genomics; 3 , 22-32 . PubMedID: 20161833
- Gautier A. et al. (2009) Selective cross-linking of interacting proteins using self-labeling tags J Am Chem Soc; 131, 17954-17962 . PubMedID: 19916541
- Donovan C. et al. (2009) Characterization and subcellular localization of bacterial flotillin homologue Microbiology; 155 , 1786-1799 . PubMedID: 19383680
- Cornish, V. W. (2009) Fluorescence in living systems: applications in chemical biology Wiley Encyc. of Chem. Biol. ; 2 , 28-38 .
- Kapmeier F. et al. (2009) Site-Specific, covalent labeling of recombinant antibody fragments via fusion to an engineered version of 6-O-alkylguanine DNA alkyltransferase Bioconjugate Chem; 23-Apr , PubMedID: 19388673
- Gralle M. et al. (2009) Neuroprotective secreted amyloid precursor protein acts by disrupting amyloid precursor protein dimers J Biol Chem; 284, 15016-15025 . PubMedID: 19336403
- Stein V. and Hollfeder F. (2009) An efficient method to assemble linear DNA templates for in vitro screening and selection systems Nucleic Acids Res; 37, e122/1-e122/9 . PubMedID: 19617373
- Hill Z. B. (2009) A chemical genetic method for generating bivalent inhibitors of protein kinases J Am Chem Soc; 131, 6686-6688 . PubMedID: 19391594
- Johnsson K. (2009) Visualizing biochemical activities in living cells Nat Chem Biol; 5 , 63-65 . PubMedID: 19148167
- Carroll C.W. et al. (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N Nat�Cell�Biol; 11 , 896-902 . PubMedID: 19543270
- Samoshkin A. et al. (2009) Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation PLoS One; 4 , e6831 . PubMedID: 19714251
- Keppler A. et al. (2009) Chromophore-assisted laser inactivation of α- and γ-tubulin SNAP-tag fusion proteins inside living cells ACS Chem Biol; 4 , 127-138 . PubMedID: 19191588
- Maffei, M., Morelli, C., Graham, E., Patriarca, S., Donzelli, L., Doleschall, B., de Castro, Reis, F., Nocchi, L., Chadick, C.H., Reymond, L., Correa, I.R., Jr., Johnsson, K., Hackett, J.A., Heppenstall, P.A (2019) A ligand based system for receptor specific delivery of proteins Sci Rep; 9(1), 19214.. PubMedID: 31844114, DOI: 10.1038/s41598-019-55797-1
- Simultaneous dual protein labeling inside live cells
- Protein localization and translocation
- Pulse-chase experiments
- Receptor internalization studies
- Selective cell surface labeling
- Protein pull-down assays
- Protein detection in SDS-PAGE
- Flow cytometry
- High throughput binding assays in microtiter plates
- Biosensor interaction experiments
- FRET-based binding assays
- Single molecule labeling
- Super-resolution microscopy
Lukinavičius, G. et al. (2015) "Fluorescent labeling of SNAP-tagged proteins in cells" Methods Mol. Biol. 1266, 107-118.
Corrêa Jr., I. R. (2015) "Considerations and protocols for the synthesis of custom protein labeling probes" Methods Mol. Biol. 1266, 55-79.
Corrêa Jr., I. R. (2014) "Live-cell reporters for fluorescence imaging" Curr. Opin. Chem. Biol. 20, 36-45.
Single-Molecule Imaging:
Bosch, P. J. et al. (2014) "Evaluation of fluorophores to label SNAP-tag fused proteins for multicolor single-molecule tracking microscopy in live cells" Biophys. J. 107, 803-814.
Smith, B. A. et al. (2013) "Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation" eLife 2, e01008.
Jaiswal, R. et al. (2013) "The Formin Daam1 and Fascin Directly Collaborate to Promote Filopodia Formation" Curr. Biol. 23, 1373-1379.
Breitsprecher, D. et al. (2012) "Rocket Launcher Mechanism of Collaborative Actin Assembly Defined by Single-Molecule Imaging" Science 336, 1164-1168.
Hoskins, A. A. et al. (2011) "Ordered and dynamic assembly of single spliceosomes." Science 331 (6022), 1289-1295.
Super-Resolution Imaging:
Zhao, Z. W. et al. (2014) "Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy" Proc. Natl. Acad. Sci. USA 111, 681-686.
Lukinavičius, G. et al. (2013) "A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins" Nat. Chem. 5, 132-139.
Jones, S. A. et al. (2011) "Fast, three-dimensional super-resolution imaging of live cells." Nat. Methods 8, 499-505.
Klein, T. et al. (2011) "Live-cell dSTORM with SNAP-tag fusion proteins." Nat. Methods 8, 7-9.
Pellett, P. A. et al. (2011) "Two-color STED microscopy in living cells." Biomed. Opt. Expr. 2, 2364-2371
Hein, B. et al. (2010) "Stimulated Emission Depletion Nanoscopy of Living Cells Using SNAP-Tag Fusion Proteins." Biophys. J. 98, 158-163.
Tissue and Animal Imaging:
Yang, G. et al. (2015) "Genetic targeting of chemical indicators in vivo" Nat. Methods 12, 137-139.
Kohl, J. et al. (2014) "Ultrafast tissue staining with chemical tags" Proc. Natl. Acad. Sci. USA 111, E3805-E3814.
Ivanova, A. et al. (2013) "Age-dependent labeling and imaging of insulin secretory granules" Diabetes 62, 3687-3696.
Gong, H. et al. (2012) "Near-Infrared Fluorescence Imaging of Mammalian Cells and Xenograft Tumors with SNAP-Tag" PLoS ONE 7(3): e34003.
Bojkowska K. et al. (2011) "Measuring in vivo protein half-life." Chem. Biol. 18, 805-815.
Cell-Surface Protein Labeling and Internalization Analysis:
Bitsikas, V. et al. (2014) "Clathrin-independent pathways do not contribute significantly to endocytic flux" eLife 3, e03970.
Jaensch, N. et al. (2014) "Stable Cell Surface Expression of GPI-Anchored Proteins, but not Intracellular Transport, Depends on their Fatty Acid Structure" Traffic 15, 1305-1329.
Cole, N. B. and Donaldson, J. G. (2012) "Releasable SNAP-tag Probes for Studying Endocytosis and Recycling" ACS Chem. Biol. 7, 464-469.
Pulse-Chase Analysis:
Rošić, S. et al. (2014) "Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division" J. Cell Biol. 207, 335-349.
Stoops, E. H. et al. (2014) "SNAP-Tag to Monitor Trafficking of Membrane Proteins in Polarized Epithelial Cells" Methods Mol. Biol. 1174, 171-182.
Bordor, D. L. et al. (2012) "Analysis of Protein Turnover by Quantitative SNAP-Based Pulse-Chase Imaging" Curr. Protoc. Cell Biol. 55, 8.8.1-8.8.34.
Pull-Down Studies:
Register, A. C. et al. (2014) "SH2-Catalytic Domain Linker Heterogeneity Influences Allosteric Coupling across the SFK Family" Biochemistry 53, 6910-6923.
Shi, G. et al. (2012) "SNAP-tag based proteomics approach for the study of the retrograde route" Traffic 13, 914-925.
Bieling, P. et al. (2010) "A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps" Cell 142, 420-432.
Protein-Protein and Protein-Ligand Interactions:
Griss, R. et al. (2014) "Bioluminescent sensor proteins for point-of-care therapeutic drug monitoring" Nat. Chem. Biol. 10, 598-603.
Chidley, C. et al. (2011) "A yeast-based screen reveals that sulfasalazine inhibits tetrahydrobiopterin biosynthesis." Nat. Chem. Biol. 7, 375-383.
Gautier A. et al. (2009) "Selective Cross-Linking of Interacting Proteins using Self-Labeling Tags" J. Am. Chem. Soc. 131, 17954-17962.
Maurel D. et al. (2008) "Cell-surface protein-protein interaction analysis with time-resolved FRET and SNAP-tag technologies: application to GPCR oligomerization." Nat. Methods 5, 561-567.
- Clone and express once, then use with a variety of substrates
- Non-toxic to living cells
- Wide selection of fluorescent substrates
- Highly specific covalent labeling
- Simultaneous dual labeling
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.