Kinetics and mechanism of polynucleotide ligases
DNA ligases catalyze the formation of a phosphodiester bond between an adjacent 5’ PO4 and 3’ OH in three distinct catalytic steps: a) Self-adenylation b) Adenylyl transfer and c) Phosphodiester bond formation. We are using rapid chemical quench, stopped-flow, and bulk fluorescence measurements to determine the microscopic kinetic rates that drive each of these catalytic events in several viral, archaeal and human DNA ligases. Our work seeks to understand the mechanistic details of DNA binding, ligation site localization, conformational changes during turnover, and the chemical steps of ligation. Further, we aim to understand not only the biologically relevant nick-sealing event, but also end-joining in the absence of accessory factors, an activity critical for biotechnological ligation applications.
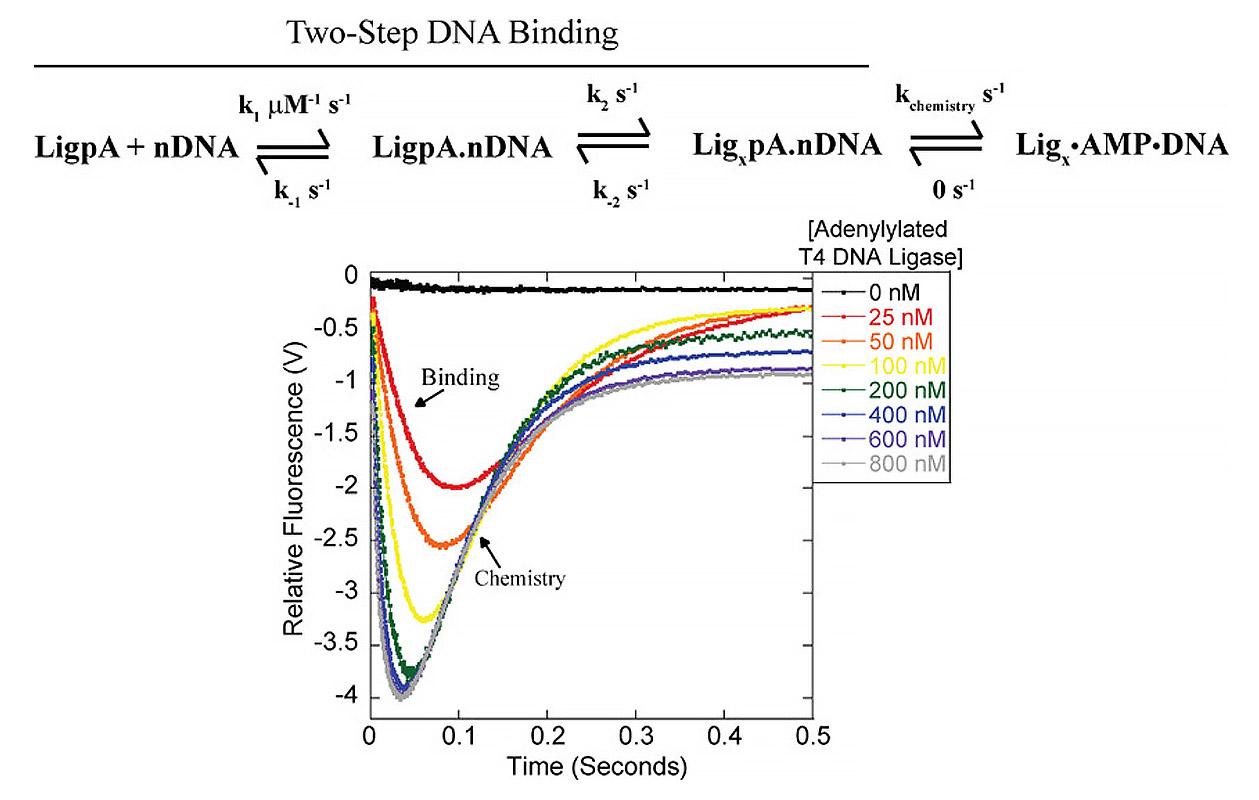
We have several ongoing collaborations aimed at broadening our approach to these questions. In collaboration with the Hoskin’s lab at University of Wisconsin- Madison, we are using fluorescently labeled DNA ligases, single-molecule FRET and bulk FRET, to investigate the real-time conformational changes of DNA ligases during the process of ligation and to probe ligase-nucleic acid binding interactions. With the Keck lab, also at University of Wisconsin- Madison, we aim to capture DNA ligase bound to DNA end-joining and other substrates.

